The Silicon Graphics Teaching Laboratory
has been replaced by the
Chemi
cal Information Laboratory
and this information is for historical interest only
Brief Introduction to the Crystallographic Database
The Centre has a WWW server
Quest no longer runs on the older workstations in the SGTL.
Consult
the upgrade information.
A Simple Search
Getting started: First decide on a name for the files which will hold the results
of the search. This name can be any number of letters and numbers, but it must not
contain any spaces. In this example, the name filename will be used. Type quest
filename.'Quest' is the command to start the searching program. A new window
will appear on the screen, which you can position with the mouse. Click the left
mouse button to acknowledge the credits, and to move to the Build menus.
Building a substructure: In this example, we will look for all the compounds in
the database which contain a certain substructure. You can draw the substructure
that you want to search for in the main window, for example, a decalin ring 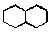 . By default, carbon atoms and single bonds are selected, but you
can change this by clicking in the appropriate places. When the substructure is
complete, click on Define-Structure. This tells the program to memorise the
structure you have drawn. If you want to search for several structures, then you can
now draw another, and click Define-Structure when it is complete. . By default, carbon atoms and single bonds are selected, but you
can change this by clicking in the appropriate places. When the substructure is
complete, click on Define-Structure. This tells the program to memorise the
structure you have drawn. If you want to search for several structures, then you can
now draw another, and click Define-Structure when it is complete.
Choose Search options: Now click on To-Search. This will enable you to
decide on the information that you want to gather. Choose REFC, ASER, FBIB, FDAT,
FCON to remember everything.
Starting the search: Now click on QUEST. This will change all the menus, and
show pictures of all the structures you have defined. There should only be one
structure, in this example, but it is possible to define several substructures or texts,
and combine them in the search with AND, OR and NOT. Click on the structure. This
will set up a search question, that will look for every structure in the database that
contains this substructure. Now click Start-Search.
During the search: Whenever the program finds a structure that contains the
substructure you defined, it will display a 2D picture of the structure, with the
substructure highlighted in red. Some structures also have a 3D picture of the
structure, which can be rotated and enlarged using the controls at the bottom right of
the screen. If the structure is one that you are interested in, then click on KEEP.
Otherwise, click REJECT.
Finishing: When the search is complete, data about the structures you kept
will be in the files whose names begin with filename. Searching for decalin will find
a very large number of structures, and you will probably want to click EXIT before
the search is complete.
Notes
- Holding down the right mouse button is equivalent to repeatedly clicking the left
mouse button.
- Text searches of various sorts are available from the SEARCH menus.
- The default rotation increment is 30 degrees. Click this number and change it to 2 degrees for
smoother rotation.
- To view a crystal structure in MacroModel, you must convert the file format. You
need to have both an FDAT file and a CONN file. These will be called filename.dat
and filename.con. Type 'csd2mmod filename' to create a new file filename.mmod.
- IsoStar is now available
|