Practical Seven: More Problems
(1) Account for the selectivity of a rearrangement
This diol undergoes a pinacol rearrangement when treated with a Lewis
acid (Et2O.BF3). The pinacol rearrangement could form a mixture of two
different ketones, but it the experiment shows that only one is formed. In
this practical you will use molecular mechanics to analyse this reaction,
and to rationalise the observed selectivity.
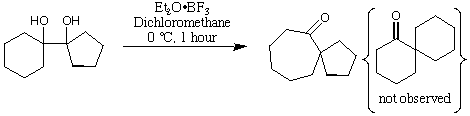 (a) Assume that the reaction is thermodynamically controlled. In this case,
the ratio of the products will be controlled by the relative energies of the
products. Calculate the relative energies of the products using
MacroModel, and the MM2 force field. You could use a Monte-Carlo
search to ensure you have a global minimum energy in each case, but it
may be quicker to do the search by hand as the ketones are not very
flexible.
(a) Assume that the reaction is thermodynamically controlled. In this case,
the ratio of the products will be controlled by the relative energies of the
products. Calculate the relative energies of the products using
MacroModel, and the MM2 force field. You could use a Monte-Carlo
search to ensure you have a global minimum energy in each case, but it
may be quicker to do the search by hand as the ketones are not very
flexible.
(b) When you have found the energies of the low energy conformations of
the two ketones, calculate the corresponding Boltzmann factors. The
relative proportions of the two ketones will be given by the ratio of their
Boltzmann factors. Is the experiment consistent with the premise that the
reaction is thermodynamically controlled?
(c) Assume that the reaction is kinetically controlled. We must estimate the
size of the energy barrier that must be overcome to form the products. In
order to do this, note that the carbonium ion formed by removal of one of
the hydroxyl groups will be similar in structure to the transition state of the
reaction. Calculate the relative energies of the two carbonium ions. What
ratio of products is expected now?
(d) Are your results consistent with the experiment?
(2) Design a reagent for a stereoselective aldol reaction
MacroModel's version of the MM2 force field (MM2*) contains parameters
that allow it to investigate the structures of the transition state of the boron-
mediated aldol reaction. The parameters are only applicable to the
reactions of ethyl ketones.
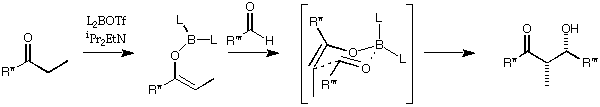 To set up the transition state, draw the following structure in MacroModel.
Two of the bonds are 'zero-order.' To draw these, make single bonds, click
once on delete, then click once in the centre of each bond. The bonds will
go green, to signify zero-order. The boron atom is a user-defined atom
type. First draw this atom as a carbon atom, then click on '*' which is just
below 'Lp' in the list of atom types. You will be asked for an atom label,
and you must type 'B3' (the capital letter is essential). Click on the carbon
atom It should go pale green, and be labelled 'B.'
To set up the transition state, draw the following structure in MacroModel.
Two of the bonds are 'zero-order.' To draw these, make single bonds, click
once on delete, then click once in the centre of each bond. The bonds will
go green, to signify zero-order. The boron atom is a user-defined atom
type. First draw this atom as a carbon atom, then click on '*' which is just
below 'Lp' in the list of atom types. You will be asked for an atom label,
and you must type 'B3' (the capital letter is essential). Click on the carbon
atom It should go pale green, and be labelled 'B.'
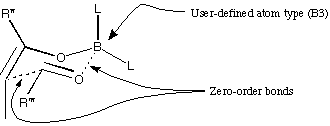 Once you have set up this transition state, you can minimise it, using the
MM2* force field. When you start a minimisation, MacroModel will suggest
that the lone pairs do not match the force field, and ask to alter them.
Usually you would ask MacroModel to alter the lone pairs for you, but in
this case, click 'No.'
Once you have set up this transition state, you can minimise it, using the
MM2* force field. When you start a minimisation, MacroModel will suggest
that the lone pairs do not match the force field, and ask to alter them.
Usually you would ask MacroModel to alter the lone pairs for you, but in
this case, click 'No.'
The energies you obtain for these structures should correspond to the
energies of the transition states of the aldol reaction. If you calculate the
energies of diastereomeric transition states, you should get a measure of
the selectivity of the reaction.
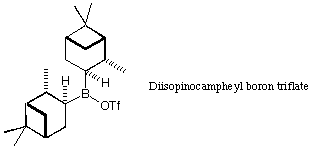 Use this tool to design a selective ligand. Ideally, you would do a full
conformational search on each, but if you choose fairly rigid structures,
you may be able to get an idea of the global minimum by adjusting the
torsion angles by hand (use the Rot T button). The
diisopinocampheyl ligand has often been used as a chiral auxiliary in this
reaction. Can you design something better?
Use this tool to design a selective ligand. Ideally, you would do a full
conformational search on each, but if you choose fairly rigid structures,
you may be able to get an idea of the global minimum by adjusting the
torsion angles by hand (use the Rot T button). The
diisopinocampheyl ligand has often been used as a chiral auxiliary in this
reaction. Can you design something better?
(3) Design a ligand for ristocetin
Ristocetin is an antibiotic which inhibits bacterial cell wall growth. Like
penicillin, it exploits the unusual D-ala-D-ala peptide sequence which is
present in cell wall formation. Penicillin mimics this unusual structure, but
ristocetin binds to its, thus inhibiting the growth of cell walls. The files
/usr/local/examples/rist.dat and /usr/local/examples/ristDD.dat contain the
structures of aglyco-ristocetin and acetyl-D-alanine-D-alanine complexed
to aglycoristocetin. A thorough study of the binding interactions would
require long molecular dynamics simulations, but a qualitative picture
emerges from simple minimisations. Use the AMBER force field, and
solvent models to investigate the structures.
Experimentally, acetyl-D-alanine-D-alanine binds more strongly than any
of its diastereoisomers, or the analogues in which one or both alanines is
replaced by glycine. Is this what you would expect from the model?
Experiments in vacuo show a much reduced selectivity. Can you
rationalise this? Might other peptidic or non-peptidic molecules bind as
well or better than acetyl-D-alanine-D-alanine? It will not be possible to get
quantitative answers to these questions in the time available, but it may be
possible to get a qualitative picture of the likely interactions.
(4) Investigate Enzyme Structure
The structures of several enzymes are in the
/usr/local/examples/enzymes. These are an aldolase, carboxypeptidase A
bound to an inhibitor, lysozyme bound to a trisaccharide (the natural
substrate is a tetrasaccharide), an RNase, and triosephosphate
isomerase.
MacroModel can read each of these structures, and manipulate them.
They are too big to minimise directly, but you can use the SUBED
submode to select the area around the active site for minimisation, or for
molecular dynamics. What other molecules might inhibit these enzymes?
|