The Silicon Graphics Teaching Laboratory
has been replaced by the
Chemi
cal Information Laboratory
and this information is for historical interest only
Computational Biological Chemistry
The Silicon Graphics Teaching Laboratory provides a wide range of programs, including many that are
specifically intended for the study of biological chemistry. This page outlines
the programs which were written with biological applications in mind.
A complete
list of the programs licensed in the laboratory is also available.
A few of the facilities available are illustrated in the following thumbnail pictures,
all of which were created using programs in the laboratory.
Click on any of the small pictures to see a larger version and a brief explanation.
The pictures illustrate methods of investigating peptides, drug-design, explicit and
implicit solvent models, and molecular dynamics studies of oligosaccharides.
 |
 |
 |
| Eadfrith | MacroModel | Cerius |
| | |
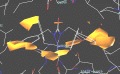 |
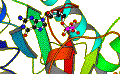 |
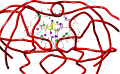 |
| Sybyl/Grid | Molscript | Xed/Cosmic |
| | |
 |
 |
 |
Oligosaccharides
Continuum Solvent | Oligosaccharides
Dynamics | Ligand Binding
Explicit Solvent |
Programs Available
Advice on the use of all the following programs is available online.
If the instructions need to be expanded or corrected, please E-mail detailed suggestions
to jmg11@cam.ac.uk.
Molecular Modelling Programs
- These programs are able to do almost any sort of biological chemistry modelling.
Insight is the only major program which is not available at the moment.
However, MSI intend to merge Insight with Cerius2 as soon as possible.
- MacroModel 5.5
- Sybyl
- Cerius2
- Xed/Cosmic
Databases
Drug Design
- Dock
A program to design receptors for enzyme active sites
- GASP Genetic Algorithms to align molecules and so aid in the identification of
a pharmacophore (consult Jack Bikker, jab44@cam.ac.uk, http://mrical.ch.cam.ac.uk:8090/)
- Grid: Determines energetically favourable binding sites for ligands
(Documentation is only available from SGTL)
- Delphi A program to calculate, display and analyse electrostatic potentials of macromolecules
Molecular Graphics
Miscellaneous
- ANSIG Interpretation of NMR spectra
(Documentation is only available from SGTL)
- Babel
A program to convert between different molecular modelling
file formats.
|