
- Documentation as PDF files in the directory: /usr/glea/schrodinger/docs/maestro5/pdf/
Maestro is a graphical user interface to both
MacroModel and
Jaguar. It is started by typing:
maestro
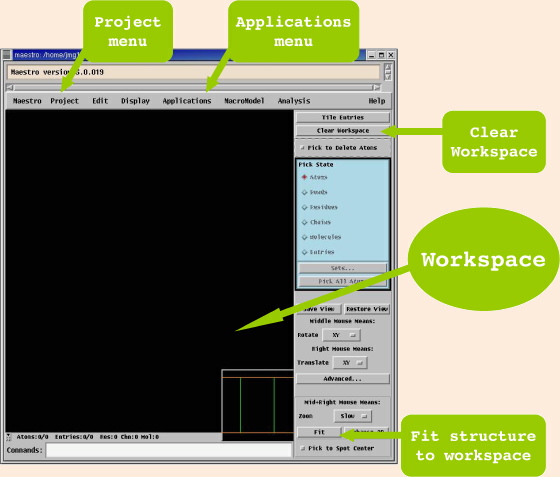
This command creates a window, most of which is the black area
of the Workspace, with menus along the top, and other
options to the right.
Maestro organises molecules into projects. The first
thing to do is to select a project, either by making a new
one (Menu: Project->New Project...) or by opening an
existing one (Menu: Project->Open Project...). This
will create a Project Table which can be used to keep
track of a list of molecules, each of which is an entry
in the project. The Project Table does not save the structures -
if a molecule appears in the Project Table it is possible that
the files describing the molecule have already been deleted. Usually you
will do a calculation on a molecule after building it, and this will
save the structure as a file.
Molecules are imported into Maestro either by importing them
from a file (Menu: Project->Import Structure...) or by building them
from scratch (Menu: Project->Build...). Both these options
creates a new panel, with a variety of options: the first for
selecting files of molecules, and controlling how many molecules
are imported from it; the second with a wide range of options for
drawing molecules.
A molecule in the Workspace can be rotated by holding down the
middle mouse button (or mouse wheel) and moving the mouse.
They can be translated or re-centred using the right-hand mouse
button. They can be zoomed by pressing the middle and right
buttons together.
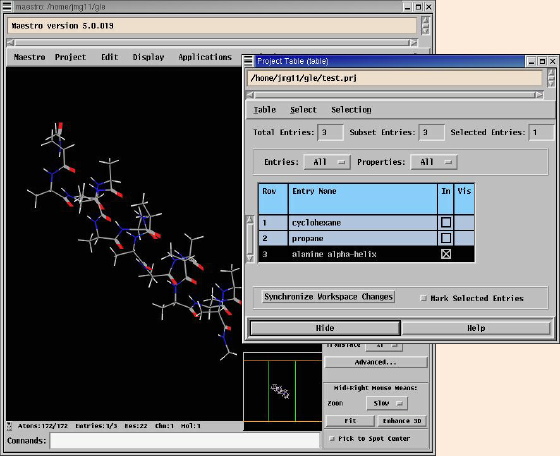
Molecules which are imported from files are automatically added
to the Project Table. The third column of this contains
checkboxes, which can be selected to decide which molecules from the
project will be shown on the Workspace. In the picture below
the Project Table lists cyclohexane, propane and an
alanine alpha-helix, the last of which is selected in the
third column of the table, and so displayed in the Workspace.
Molecules which are built in the Workspace are not put in the
Project Table unless the computer is asked to do this
(Menu: Project->Create Entry from Workspace...).
This does not record the structure. To do this, you should either
do a calculation on the structure, or else (Menu: Project->Export...)
the structure to a file.
Quitting from Maestro (Menu: Maestro->Quit...; You have the option
of saving the log file of the session, but the default is to delete it) loses the
molecules in the Workspace. You can keep them for another session by
exporting them to a file (Menu: Project->Export...). However,
molecules are usually drawn so that calculations can be done on them, and
both MacroModel and
Jaguar will automatically create files to
keep track of the calculations underway.
Once a molecule has been drawn, choose either MacroModel or
Jaguar from the menu Applications.
These will enable you to do calculations on the molecule.
|